

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
Absolute and Difference solversGet data from the RPI tank phantomThis tutorial is based on the tank phantom data contributed by Jon Newell. Data were measured on a cylindrical tank shown below. The data can by assumed to be 2D because the vertical dimension is constant.
Figure: Figure: Phantom Image (from Isaacson, Mueller, Newell and Siltanen, IEEE Trans Med Imaging 23(7): 821-828, 2004) Build a good FEM of the phantomIt is important to correctly model the size of the electrodes and their position to get absolute imaging to work.
% RPI tank model $Id: rpi_data01.m 3790 2013-04-04 15:41:27Z aadler $
Nel= 32; %Number of elecs
Zc = .0001; % Contact impedance
curr = 20; % applied current mA
th= linspace(0,360,Nel+1)';th(1)=[];
els = [90-th]*[1,0]; % [radius (clockwise), z=0]
elec_sz = 1/6;
fmdl= ng_mk_cyl_models([0,1,0.1],els,[elec_sz,0,0.03]);
for i=1:Nel
fmdl.electrode(i).z_contact= Zc;
end
% Trig stim patterns
th= linspace(0,2*pi,Nel+1)';th(1)=[];
for i=1:Nel-1;
if i<=Nel/2;
stim(i).stim_pattern = curr*cos(th*i);
else;
stim(i).stim_pattern = curr*sin(th*( i - Nel/2 ));
end
stim(i).meas_pattern= eye(Nel)-ones(Nel)/Nel;
stim(i).stimulation = 'Amp';
end
fmdl.stimulation = stim;
clf; show_fem(fmdl,[0,1])
print_convert('rpi_data01a.png','-density 60');
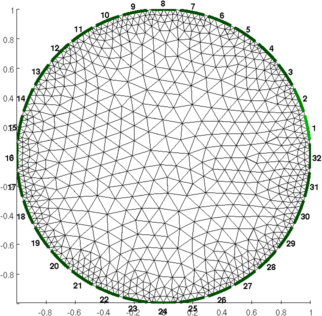
Figure: Figure: FEM phantom Load and preprocess dataOne can improve the equality of the images by ensuring that all channels have a mean voltage of zero before beginning.
% load RPI tank data $Id: rpi_data02a.m 2236 2010-07-04 14:16:21Z aadler $
load Rensselaer_EIT_Phantom;
vh = real(ACT2006_homog);
vi = real(ACT2000_phant);
if 1
% Preprocessing data. We ensure that each voltage sequence sums to zero
for i=0:30
idx = 32*i + (1:32);
vh(idx) = vh(idx) - mean(vh(idx));
vi(idx) = vi(idx) - mean(vi(idx));
end
end
Difference imagingUsing a very simple model
% RPI tank model $Id: rpi_data02b.m 3790 2013-04-04 15:41:27Z aadler $
% simple inverse model -> replace fields to match this model
imdl = mk_common_model('b2c2',32);
imdl.fwd_model = mdl_normalize(imdl.fwd_model, 0);
imdl.fwd_model.electrode = imdl.fwd_model.electrode([8:-1:1, 32:-1:9]);
imdl.fwd_model = rmfield(imdl.fwd_model,'meas_select');
imdl.fwd_model.stimulation = stim;
imdl.hyperparameter.value = 1.00;
% Reconstruct image
img = inv_solve(imdl, vh, vi);
img.calc_colours.cb_shrink_move = [0.5,0.8,0.02];
img.calc_colours.ref_level = 0;
clf; show_fem(img,[1,1]); axis off; axis image
print_convert('rpi_data02a.png','-density 60');
Using a the accurate electrode model
% RPI tank model $Id: rpi_data03.m 3790 2013-04-04 15:41:27Z aadler $
% simple inverse model -> replace fields to match this model
imdl.fwd_model = fmdl;
img = inv_solve(imdl , vh, vi);
img.calc_colours.cb_shrink_move = [0.5,0.8,0.02];
img.calc_colours.ref_level = 0;
clf; show_fem(img,[1,1]); axis off; axis image
print_convert('rpi_data03a.png','-density 60');
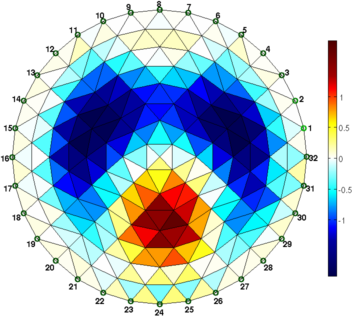
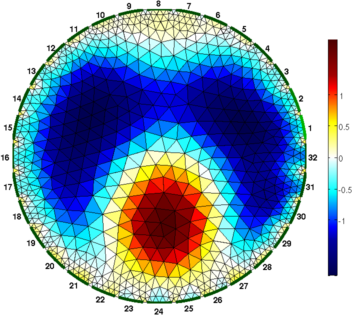
Figure: Figure: Differences images reconstructed of the phantom. For difference imaging, the simple model works surprisingly well. Estimating actual conductivitiesIn order to estimate the actual conductivity values, we need to scale for the applied voltage, tank size (in 3D) and background conductivity
% RPI tank model $Id: rpi_data04.m 5509 2017-06-06 14:33:29Z aadler $
% In 3D, it's important to get the model diameter right, 2D is
imdl.fwd_model.nodes= imdl.fwd_model.nodes*15; % 30 cm diameter
% Estimate the background conductivity
imgs = mk_image(imdl);
vs = fwd_solve(imgs); vs = vs.meas;
pf = polyfit(vh,vs,1);
imdl.jacobian_bkgnd.value = pf(1)*imdl.jacobian_bkgnd.value;
imdl.hyperparameter.value = imdl.hyperparameter.value/pf(1)^2;
img = inv_solve(imdl, vh, vi);
img.calc_colours.cb_shrink_move = [0.5,0.8,0.02];
img.calc_colours.ref_level = 0;
clf; show_fem(img,[1,1]); axis off; axis image
print_convert('rpi_data04a.png','-density 60');
This can even be a quick way to use a difference solver
for absolute imaging
% Cheap static solver $Id: rpi_data05.m 3790 2013-04-04 15:41:27Z aadler $
% Do a diff solve with respect to simulated data
imgs = mk_image(imdl);
vs = fwd_solve(imgs); vs = vs.meas;
imdl.hyperparameter.value = 1.00;
img = inv_solve(imdl, vs, vi);
img.elem_data = pf(1) + img.elem_data*0.5;
img.calc_colours.cb_shrink_move = [0.5,0.8,0.02];
clf; show_fem(img,[1,1]); axis off; axis image
print_convert('rpi_data05a.png','-density 60');
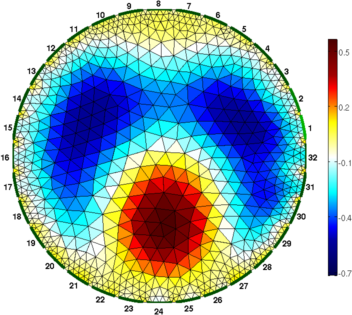
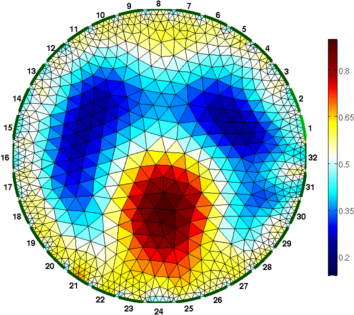
Figure: Difference (right) and one step absolute (left) images Absolute solvers as a function of iteration
% Absolute reconstructions $Id: rpi_data06.m 5537 2017-06-14 12:49:20Z aadler $
imdl = mk_common_model('b2c2',32);
imdl.fwd_model = fmdl;
imdl.reconst_type = 'absolute';
imdl.hyperparameter.value = 2.0;
imdl.solve = @inv_solve_abs_GN;
for iter = [1,2,3, 5];
imdl.inv_solve_gn.max_iterations = iter;
img = inv_solve(imdl , vi);
img.calc_colours.cb_shrink_move = [0.5,0.8,0.02];
img.calc_colours.ref_level = 0.6;
clf; show_fem(img,[1,1]); axis off; axis image
print_convert(sprintf('rpi_data06%c.png', 'a'-1+iter),'-density 60');
end
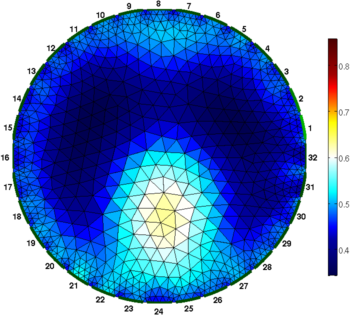
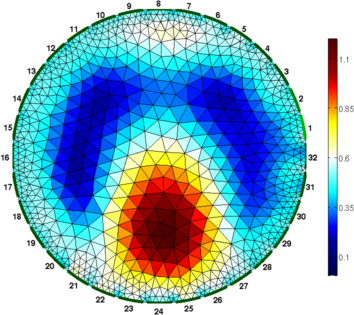
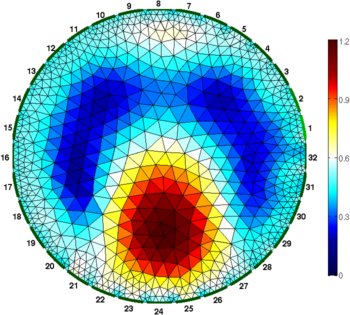
Figure: Gauss-Newton Absolute solver on conductivity at iterations (from left to right): 1, 3, 5
% Absolute reconstructions $Id: rpi_data07.m 4986 2015-05-11 20:09:28Z aadler $
imdl = mk_common_model('b2c2',32);
imdl.fwd_model = fmdl;
imdl.reconst_type = 'absolute';
imdl.hyperparameter.value = 2.0;
imdl.solve = @inv_solve_abs_CG;
imdl.inv_solve_abs_CG.elem_working = 'log_conductivity';
imdl.inv_solve_abs_CG.elem_output = 'conductivity';
for iter = [1,2,3, 5];
imdl.parameters.max_iterations = iter;
img = inv_solve(imdl , vi);
img.calc_colours.cb_shrink_move = [0.5,0.8,0.02];
img.calc_colours.ref_level = 0.6;
clf; show_fem(img,[1,1]); axis off; axis image
print_convert(sprintf('rpi_data07%c.png', 'a'-1+iter),'-density 60');
end
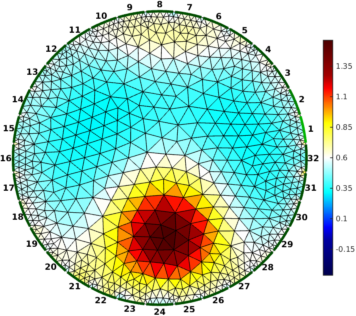
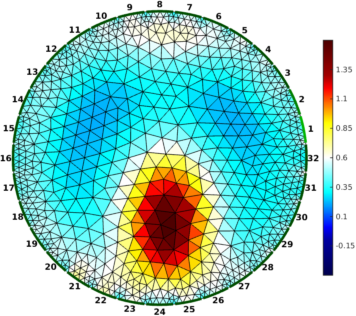
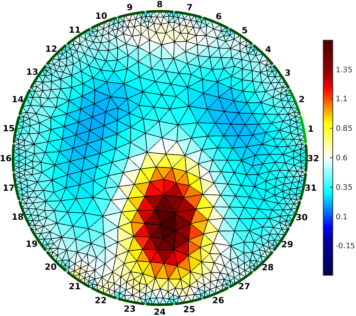
Figure: Conjugate-Gradient Absolute solver on log conductivity at iterations (from left to right): 1, 3, 5 |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $