

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
Using EIDORS to image gastric emptying2D EIT for imaging of gastric emptyingThese data were gathered by asking volunteers to drink Bovril (salty soup mix) and taking EIT images at the level of the stomach. These experiments were part of
Step 1: Create modelsUse mk_common_model to create a thorax shaped model with 16 electrodes. Ensure the model uses 1) Correct stimulation patterns (adjacent is default), 2) Normalized difference imaging
% Lung images
% $Id: tutorial410a.m 3340 2012-07-01 21:25:30Z bgrychtol $
% 2D Model
imdl= mk_common_model('c2t3',16);
% most EIT systems image best with normalized difference
imdl.fwd_model = mdl_normalize(imdl.fwd_model, 1);
imdl.RtR_prior= @prior_gaussian_HPF;
% electrodes start on back (dorsal), then do this
imdl.fwd_model.electrode([9:16,1:8])= ...
imdl.fwd_model.electrode;
subplot(221);
show_fem(imdl.fwd_model);
axis equal
print_convert tutorial410a.png;
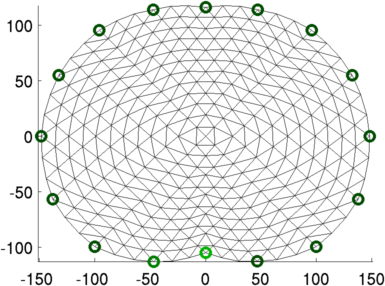
Figure: 2D FEM of thorax (units in mm). Electrode #1 (o) and electrode #2 (o) are shown in different colours than the others (o). Image reconstruction% Abdomen Images $Id: tutorial410b.m 4839 2015-03-30 07:44:50Z aadler $ load montreal_data_1995 imdl.hyperparameter.value=.2; vh= zc_h_stomach_pre; % abdomen before fluid vi= zc_stomach_0_5_60min; % each 5 minutes after drink img= inv_solve(imdl, vh, vi); clf; show_slices(img) axis equal print_convert tutorial410b.png; 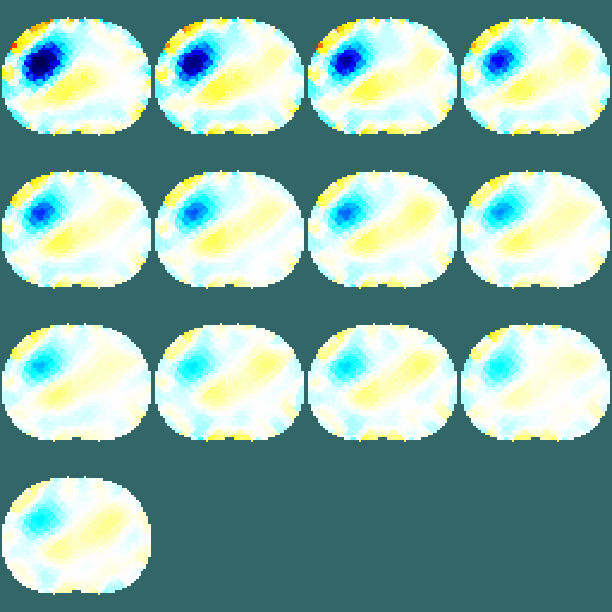
Figure: Image slices of the abdomen every five minutes after drink. Image progression is from left to right, top to bottom. Calculate signal as a function of time
% Abdomen Images $Id: tutorial410c.m 3273 2012-06-30 18:00:35Z aadler $
raster_img= calc_slices(img);
raster_img(isnan(raster_img))=0;
% define roi as whole image
s_ri = size(raster_img);
roi = ones(s_ri(1:2));
for i= 1:s_ri(3)
sig(i)= sum(sum(raster_img(:,:,i) .* roi));
end
subplot(221)
plot( ((1:s_ri(3))-1)*5, sig/sig(1))
xlabel('minutes after drink')
ylabel('normalized conductivity');
print_convert tutorial410c.png '-density 150';
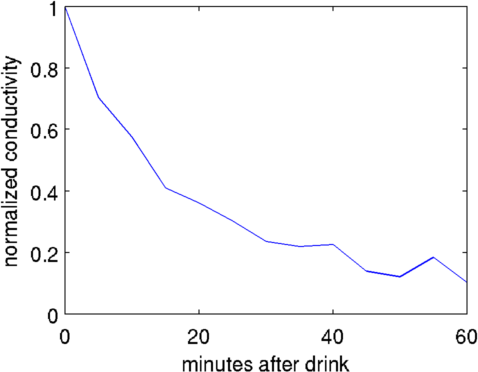
Figure: Normalized image in stomach as a function of time. Different algorithms for stomach images% Abdomen Images $Id: tutorial410d.m 4839 2015-03-30 07:44:50Z aadler $ load montreal_data_1995 vh= zc_h_stomach_pre; % abdomen before fluid vi= zc_stomach_0_5_60min(:,1); % right after drink % GN solution - Gaussian prior imdl.RtR_prior= @prior_gaussian_HPF; imdl.solve= @inv_solve_diff_GN_one_step; imdl.hyperparameter.value=.1; img= inv_solve(imdl, vh, vi); subplot(221); show_fem(img); axis equal; axis off % GN solution - Noser prior imdl.RtR_prior= @prior_noser; imdl.solve= @inv_solve_diff_GN_one_step; imdl.hyperparameter.value=.3; img= inv_solve(imdl, vh, vi); subplot(222); show_fem(img); axis equal; axis off % GN solution - Noser prior imdl= rmfield(imdl,'RtR_prior'); imdl.R_prior= @prior_TV; imdl.solve= @inv_solve_TV_pdipm; imdl.hyperparameter.value=3e-4; img= inv_solve(imdl, vh, vi); subplot(223); show_fem(img); axis equal; axis off % GN solution - Noser prior imdl.R_prior= @prior_TV; imdl.solve= @inv_solve_TV_pdipm; imdl.hyperparameter.value=3e-3; img= inv_solve(imdl, vh, vi); subplot(224); show_fem(img); axis equal; axis off print_convert tutorial410d.png '-density 150'; 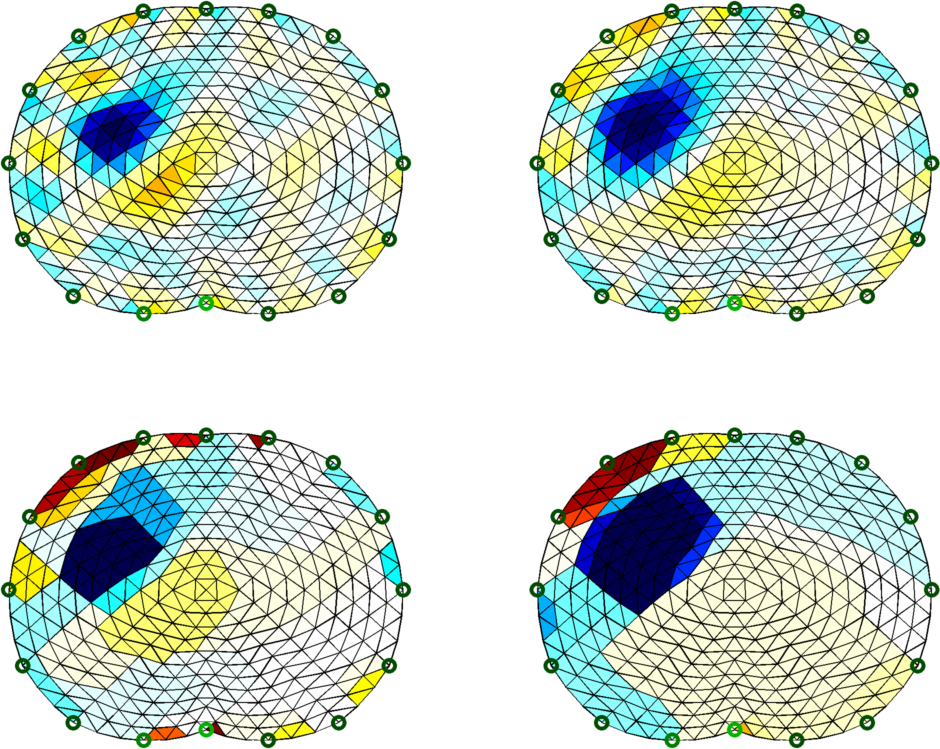
Figure: Top Left: Gauss-Newton Reconstruction with Gaussian HPF prior Top Right: Gauss-Newton Reconstruction with Laplacian filter Bottom Left: Total Variation Reconstruction (hp=1e-3) Bottom Right: Total Variation Reconstruction (hp=1e-4) |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $