

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
Introduction to dual modelsDual model systems use two models of the underlying system:
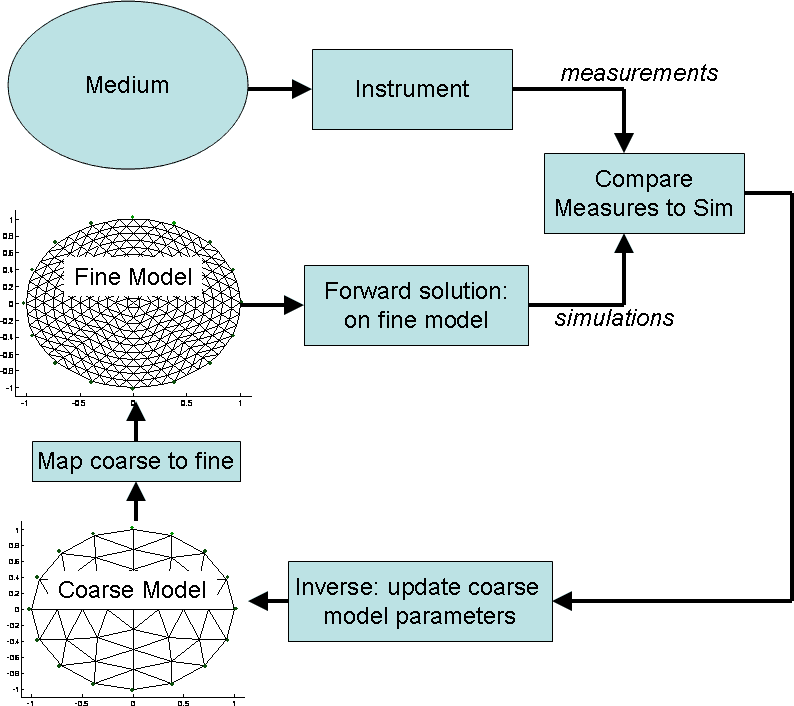
Figure: Schematic of a dual model based inverse problem Simulated dataFirst, simulate some data, based on a 576 element mesh
% Simulate data $Id: dual_model01.m 2162 2010-04-04 20:49:45Z aadler $
imdl= mk_common_model('c2c2',16);
img= mk_image(imdl);
vh=fwd_solve(img);
idx=[365,328,292,259,227,198,170,145,121];
img.elem_data([idx,idx+1,101,81])=1.1;
vi=fwd_solve(img);
show_fem(img);
print_convert('dual_model01a.png', '-density 60');
Figure: Simulation data Simple Example: Dual model using multi-element parametersNext, we compare two coarse meshes
% Simulate data $Id: dual_model02.m 4839 2015-03-30 07:44:50Z aadler $
% create base model
mdl_base=mk_common_model('a2c0',16);
mdl_base.RtR_prior = @prior_noser;
mdl_base.hyperparameter.value = .2;
elems= mdl_base.fwd_model.elems;
nodes= mdl_base.fwd_model.nodes;
e= size(elems,1);
for model = 1:3
if model==1
% Model 1: coarse==fine. each elem has a parameter
params= 1:e;
elseif model==2
% Model 2: coarse model, inner circle has one parameter
params= [1,1,1,1, 2:e-3];
elseif model==3
% Model 3: coarse model, top left slice has one parameter
params= 1:e;
params([4,8,15:16,23:24,34:36])= 0;
[jnk1,jnk2,params]= unique(params);
end
% Create inverse_model
imdl(model)= mdl_base;
imdl(model).fwd_model.coarse2fine = sparse(1:e,params,1,e,max(params));
subplot(2,3, model)
show_fem(imdl(model).fwd_model);
% Show parameter numbers
numeros= reshape(sprintf('%2d',params),2,e)';
xc=mean(reshape(nodes(elems,1),e,3),2);
yc=mean(reshape(nodes(elems,2),e,3),2);
text(xc,yc,numeros,'FontSize',8, ...
'HorizontalAlignment','center');
end
print_convert dual_model02a.png
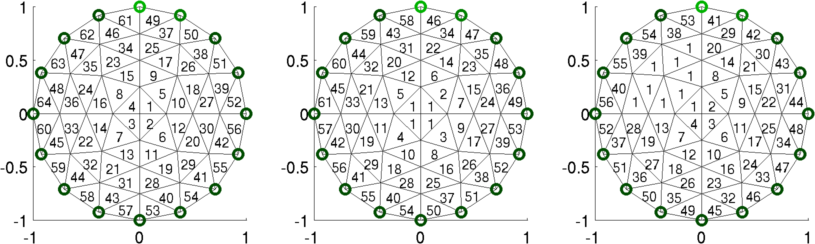
Figure: Left "Fine" mesh (64 parameters) Middle Coarse mesh (61 parameters) Right Coarse mesh (56 parameters) Coarse to Fine MappingIf we consider a smaller model, where% fine_param_1 == coarse_param_1 % fine_param_2 == coarse_param_2 % fine_param_3 == coarse_param_3 % fine_param_4 == coarse_param_3Then the mapping from coarse to fine would be [ 1 0 0 ] [ coarse_param_1 ] [ fine_param_1 ] [ 0 1 0 ] [ coarse_param_2 ] = [ fine_param_2 ] [ 0 0 1 ] [ coarse_param_3 ] [ fine_param_3 ] [ 0 0 1 ] [ fine_param_4 ] Image Reconstructions% Simulate data $Id: dual_model03.m 2162 2010-04-04 20:49:45Z aadler $ for model= 1:3 img= inv_solve(imdl(model), vh, vi); subplot(2,3,model) show_fem(img); end print_convert dual_model03a.png; 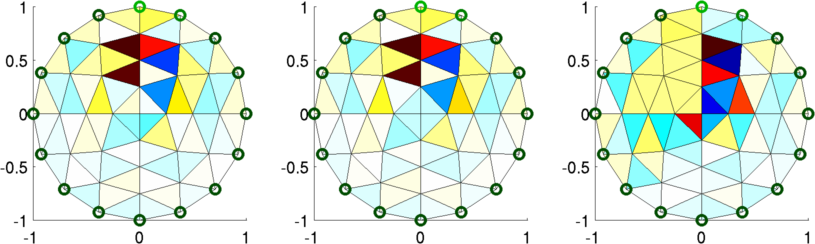
Figure: Reconstructed images. Left "Fine" mesh (64 parameters) Middle Coarse mesh (61 parameters) Right Coarse mesh (56 parameters) Example #2: Fine and Coarse meshesWe consider a coarse mesh of 64 elements, and fine meshes of 64 (same as coarse), 256, and 576 elements
% create fine meshes $Id: dual_model04.m 4839 2015-03-30 07:44:50Z aadler $
% create base model
mdl_coarse=mk_common_model('a2c0',16);
for model = 1:3
if model==1
% Model 1: 64 elements
mdl_str= 'a2c0';
elseif model==2
% Model 2: 256 elements
mdl_str= 'b2c0';
elseif model==3
% Model 3: 576 elements
mdl_str= 'c2c0';
end
mdl_fine= mk_common_model(mdl_str,16);
mdl_fine.fwd_model.mk_coarse_fine_mapping.n_interp= 150;
mdl_fine.RtR_prior = @prior_noser;
mdl_fine.hyperparameter.value = .2;
imdl(model)= mdl_fine;
imdl(model).fwd_model.coarse2fine = ...
mk_coarse_fine_mapping( mdl_fine.fwd_model, mdl_coarse.fwd_model);
subplot(2,3, model)
show_fem(mdl_fine.fwd_model);
end
print_convert dual_model04a.png
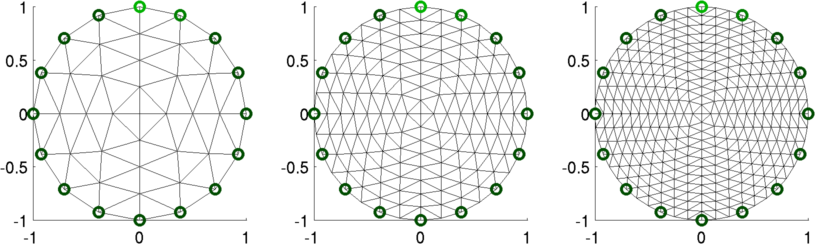
Figure: Reconstructed images. Left "Fine" mesh (64 elements) Middle Fine mesh (256 elements) Right Fine mesh (576 elements) % Simulate data $Id: dual_model05.m 2162 2010-04-04 20:49:45Z aadler $ % Reconstruct for model= 1:3 img(model)= inv_solve(imdl(model), vh, vi); end % Show image mapped to fine model for model= 1:3 subplot(2,3,model) show_fem(img(model)); end print_convert dual_model05b.png; % Show image mapped to coarse model for model= 1:3 subplot(2,3,model) img(model).fwd_model = mdl_coarse.fwd_model; show_fem(img(model)); end print_convert dual_model05a.png; 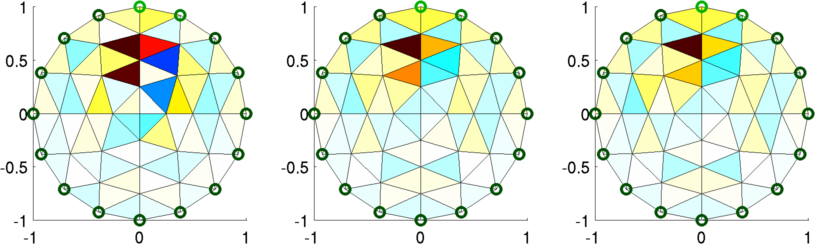
Figure: Reconstructed images mapped onto the coarse model. Left "Fine" mesh (64 elements) Middle Fine mesh (256 elements) Right Fine mesh (576 elements) 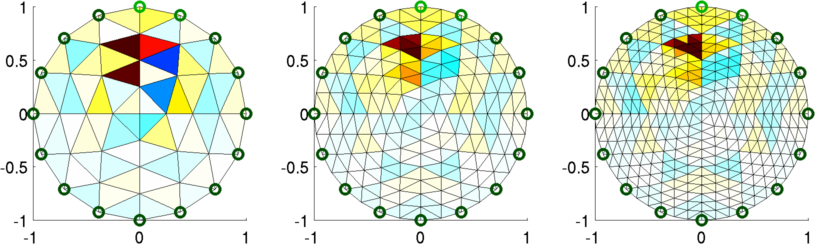
Figure: Reconstructed images mapped onto the fine model. Left "Fine" mesh (64 elements) Middle Fine mesh (256 elements) Right Fine mesh (576 elements) |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $