

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
GREIT software frameworkThe GREIT software framework is designed to simplify the analysis and allow contributions from different EIT software code bases. As shown, there are four steps
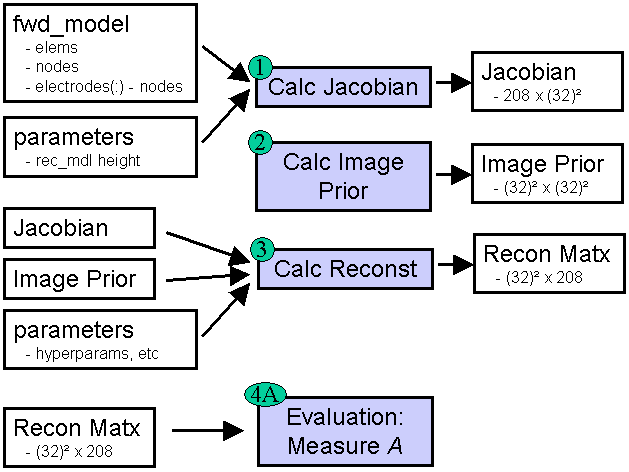
Figure: Conceptual framework for GREIT software Step 0: forward model
% fwd_model $Id: framework00.m 2684 2011-07-13 08:43:58Z aadler $
fmdl = mk_library_model('cylinder_16x1el_fine');
fmdl.stimulation = mk_stim_patterns(16,1,'{ad}','{ad}',{},1);
subplot(121)
show_fem(fmdl);
view([-5 28]);
crop_model(gca, inline('x+2*z>20','x','y','z'))
subplot(122)
show_fem(fmdl);
view(0,0);
crop_model(gca, inline('y>-10','x','y','z'))
set(gca,'Xlim',[-4,4],'Zlim',[-2,2]+5);
print_convert framework00a.png
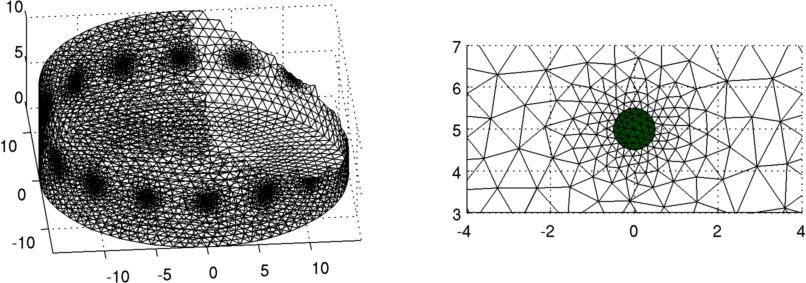
Figure: FEM model of a ring of electrodes as well. Right Refinement of mesh near the electrode. Step 1: calculate JacobianCreate a 32×32 pixel grid mesh for the inverse model, and show the correspondence between it and the find forward model.
% fwd_model $Id: framework01.m 2684 2011-07-13 08:43:58Z aadler $
fmdl = mk_library_model('cylinder_16x1el_fine');
pixel_grid= 32;
nodes= fmdl.nodes;
xyzmin= min(nodes,[],1); xyzmax= max(nodes,[],1);
xvec= linspace( xyzmin(1), xyzmax(1), pixel_grid+1);
yvec= linspace( xyzmin(2), xyzmax(2), pixel_grid+1);
zvec= [0.6*xyzmin(3)+0.4*xyzmax(3), 0.4*xyzmin(3)+0.6*xyzmax(3)];
% CALCULATE MODEL CORRESPONDENCES
[rmdl,c2f] = mk_grid_model(fmdl, xvec, yvec, zvec);
% SHOW MODEL CORRESPONDENCE
clf;
show_fem(fmdl); % fine model
crop_model(gca, inline('x-z<-8','x','y','z'))
hold on
hh=show_fem(rmdl); set(hh,'EdgeColor',[0,0,1]);
hold off
view(-47,18);
print_convert framework01a.png
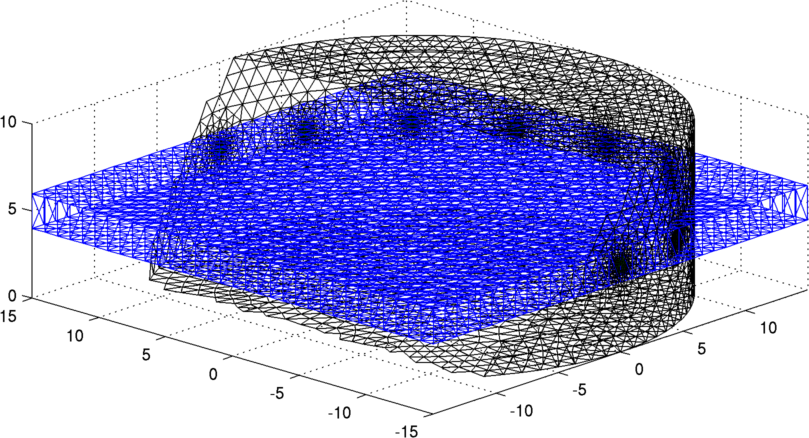
Figure: 3D fine cylindrical mesh and 32×32 reconstruction grid
% fwd_model $Id: framework02.m 3273 2012-06-30 18:00:35Z aadler $
% CALCULATE JACOBIAN AND SAVE IT
img= mk_image(fmdl, 1);
img.fwd_model.coarse2fine = c2f;
img.rec_model= rmdl;
% ADJACENT STIMULATION PATTERNS
img.fwd_model.stimulation= mk_stim_patterns(16, 1, ...
[0,1],[0,1], {'do_redundant', 'no_meas_current'}, 1);
% SOLVERS
img.fwd_model.system_mat= @system_mat_1st_order;
img.fwd_model.solve= @fwd_solve_1st_order;
img.fwd_model.jacobian= @jacobian_adjoint;
J= calc_jacobian(img);
map = reshape(sum(c2f,1),pixel_grid,pixel_grid)>0;
save GREIT_Jacobian_ng_mdl_fine J map
Step 2: calculate image priorHere we use a NOSER type prior with each diagonal element equal to √[JTJ]i,i. The √ is based on our experience that a power of zero pushes artefacts to the boundary, while a power of one pushes artefacts to the centre.% image prior $Id: framework03.m 1545 2008-07-26 17:33:46Z aadler $ load GREIT_Jacobian_ng_mdl_fine J map % Remove space outside FEM model J= J(:,map); % inefficient code - but for clarity diagJtJ = diag(J'*J) .^ (0.7); R= spdiags( diagJtJ,0, length(diagJtJ), length(diagJtJ)); save ImagePrior_diag_JtJ R Step 3: calculate reconstruction matrix% reconst $Id: framework04.m 1544 2008-07-26 17:23:52Z aadler $ load ImagePrior_diag_JtJ R load GREIT_Jacobian_ng_mdl_fine J map RM = zeros(size(J')); J = J(:,map); hp = .3; RM(map,:)= (J'*J + hp^2*R)\J'; save RM_framework_example RM map Step 4: test reconstruction matrixTo test this example reconstruction matrix, we use test data using the EIT system from IIRC, (KHU: Korea). A non-conductive object is moved in a circle in a saline tank. (Note this this example specifically does not use the EIDORS imaging functions of colour mapping, showing that the GREIT code is functionally separate from EIDORS)% Test RM $Id: framework05.m 3846 2013-04-16 16:24:32Z aadler $ % EXCLUDE MEASURES AT ELECTRODES [x,y]= meshgrid(1:16,1:16); idx= abs(x-y)>1 & abs(x-y)<15; % LOAD SOME TEST DATA load iirc_data_2006 v_reference= - real(v_reference(idx,:)); v_rotate = - real(v_rotate(idx,:)); load RM_framework_example RM map; clear imgs;for k=1:4; dv = v_rotate(:,k*2) - v_reference; img = reshape( RM*dv, 32,32); % reconstruction img(~map) = NaN; % background imgs(:,:,k) = img; end imagesc(reshape(imgs,32,4*32)) axis off; axis equal; colormap(gray(256)) print_convert framework05a.png '-density 75' 
Figure: Images of a non-conductive object moving in a tank. |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $