

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
EIDORS image reconstructionsEIDORS data structures: the inv_modelThe EIDORS inv_model describes all the parameters as part of image reconstruction
% Basic Image reconstruction
% $Id: tutorial110a.m 4839 2015-03-30 07:44:50Z aadler $
% Load some data
load iirc_data_2006
% grey background
calc_colours('greylev',-.1);
% Get a 2D image reconstruction model
imdl= mk_common_model('c2c');
% Set stimulation patterns. Use meas_current
% Stimulation of [1,0] (not [0,1]) is needed for this device (IIRC)
imdl.fwd_model.stimulation = mk_stim_patterns(16,1,[1,0],[0,1],{'meas_current'},1);
% Remove meas_select field because all 16x16 patterns are used
imdl.fwd_model = rmfield( imdl.fwd_model, 'meas_select');
vi= real(v_rotate(:,9))/1e4; vh= real(v_reference)/1e4;
for idx= 1:3
if idx==1
imdl.hyperparameter.value= .03;
elseif idx==2
imdl.hyperparameter.value= .05;
elseif idx==3
imdl.hyperparameter.value= .10;
end
img= inv_solve(imdl, vh, vi);
img.calc_colours.greylev = -.3;
subplot(2,3,idx);
show_slices(img);
subplot(2,3,idx+3);
z=calc_slices(img);
c=calc_colours(z,img);
h=mesh(z,c); view(-11,44);
set(h,'CDataMapping','Direct');
set(gca,{'XLim','YLim','ZLim','XTickLabel','YTickLabel'}, ...
{[1 64],[1 64],[-3.3,0.5],[],[]})
end
print_convert tutorial110a.png
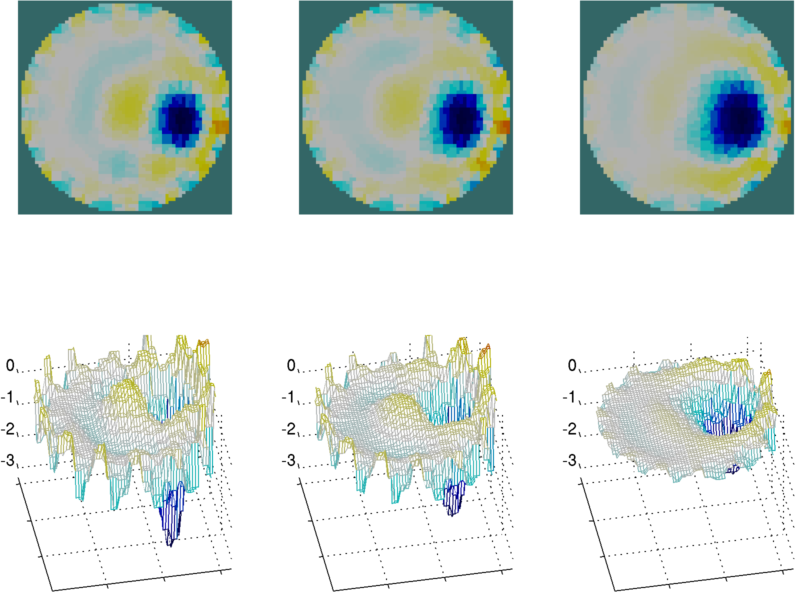
Figure: Image reconstructions shown as images (top) or meshes (bottom) for different hyperparameter values. |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $