

|
EIDORS: Electrical Impedance Tomography and Diffuse Optical Tomography Reconstruction Software |
|
EIDORS
(mirror) Main Documentation Tutorials − Image Reconst − Data Structures − Applications − FEM Modelling − GREIT − Old tutorials − Workshop Download Contrib Data GREIT Browse Docs Browse SVN News Mailing list (archive) FAQ Developer
|
One line starter program for EIDORSTo run this tutorial, you need to download and install EIDORS and then run this command in a matlab (or octave) session.run /path/to/eidors3d/startup.m Step 1: Get dataEIDORS provides lots of data to get you started. Here we choose a small file in the sample_data/ directory.Type: load montreal_data_1995 Step 2: Reconstruct and display image
show_slices(inv_solve(mk_common_model('d2c2',16),zc_h_demo4,zc_demo4))
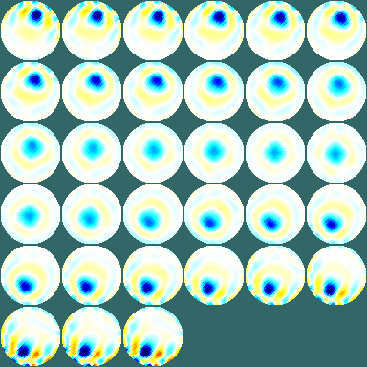
Figure: Output image showing images of a non-conductive object moving across a saline tank. One line starter program (explained in more detail)Step 1: Get dataThe key thing you need to know about your data are:
Step 2a: Create an inverse model (imdl) from the template
imdl = mk_common_model('c2c2',16);
See the documentation for mk_common_model. It has lots of options.
The function provides a circular model with adjacent stimulation patterns.
If this is not what you want, it must be changed.
Step 2b: Create a forward model (fmdl) that matches the shape / electrodesThis step is not required, if mk_common_model provides you with the shape you need. Here, as an example, we create a circular tank, but also we can use many other functions.n_rings = 12; n_electrodes = 16; three_d_layers = []; % no 3D fmdl = mk_circ_tank( n_rings , three_d_layers, n_electrodes); % then assign the fields in fmdl to imdl.fwd_model Step 2c: Create a forward model (fmdl) that matches the stimulation / measurement protocolOften the function mk_stim_patterns can do what you need; if not, you will need to:
options = {'no_meas_current','no_rotate_meas'};
[stim, meas_select] = mk_stim_patterns(16,1,'{ad}','{ad}',options,1);
imdl.fwd_model.stimulation = stim;
imdl.fwd_model.meas_select = meas_select;
If mk_stim_patterns doesn't provide what you need, then you
will need to use a function like stim_meas_list.
Step 2d: Reconstruct the image (img) using inv_solvedata_homg = zc_h_demo4; data_objs = zc_demo4; % from your file img = inv_solve(imdl, data_homg, data_objs); Step 2e: Display the imageshow_slices(img); 
Figure: Output image. |
Last Modified: $Date: 2017-02-28 13:12:08 -0500 (Tue, 28 Feb 2017) $ by $Author: aadler $