| |
ELG7173 - Final Exam Winter 2003
You have 3 hours to complete this exam.
The exam has six questions; you are required to answer any three of them.
Each question is worth equal marks.
This is a closed book exam; however,
you are permitted to bring two 8.5" × 11"
sheets of notes into the exam.
You are permitted to use a calculator.
You may not communicate with anyone during the exam except the instructor.
- Display of Medical Image Data
Software for viewing of medical images typically
provides a "windowing" function. Windowing performs
a linear stretching of the image histogram, in order
to better view pixels with values in the "window".
For example, if the screen pixels have intensities
in the range 0 to 255, a windowing from
wmin
to
wmax
will convert a pixel P to a value of
255 × (P - wmin ) /
( wmax − wmin )
on the output display. Values of P above or below this range are
"cropped" to a displayed value of 255 or 0, respectively.
Windowing serves two important functions: 1) it enhances the
visibility of contrasting tissue regions, 2) it allows selection
of the tissue type to be viewed (for example, with CT images,
windowing can allow viewing of soft tissue while bone regions
are "cropped" to the maximum or minimum value)
-
Describe how "windowing" allows these goals to be
achieved
-
Describe how the visibility of contrasting regions
in an image
varies with the size of the contrasting region and the
SNR.
-
Describe how "windowing" interacts with the
variation in visibility with SNR.
Considering these effects, what image
conditions (in terms of contrast size and SNR)
benefit most from "windowing"?
- X-Ray imaging System
A simple X-ray imaging system consists of a source, an object,
and a film detector.
-
Sketch, and describe each component from the point of view of the
generation of- and interaction with X-ray photons
-
Show the convolution form of image formation for
this X-ray system. Briefly describe each parameter and how it
relates to the physical properties of the system.
-
For each system component, list the properties that
degrade image resolution, and the magnification
level of each effect.
- C.T. Image Reconstruction
Consider the geometry defined in figure Q3 for a
model of a computed tomography system. Seven hexagonal
regions, R1 to R7 are defined.
Each region is 2 cm wide between parallel faces.
Regions R1, R2, R3
R5, R6, and R7
are soft tissue,
and
region R4 is bone.
The attenuation coefficients
for these tissues are defined in table Q3:
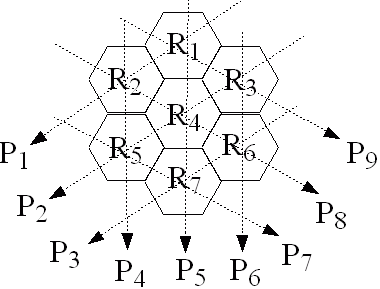
Figure Q3: Diagram for question 3.
|
|
|
| Attenuation (cm-1)
|
| Tissue
| Eγ = 20 keV
| Eγ = 50 keV
|
| Soft Tissue
| 0.805
| 0.347
|
| Bone
| 1.956
| 0.805
|
Table Q3: Tissue attenuation coefficients for question 3.
|
Along each projection,
1.39×106 X-ray photons of energy
Eγ=20keV and
1.78×105 X-ray photons of energy
Eγ=50keV
are emitted. The detectors function such that projections P
are calculated as follows:
P = log10 ∑( Nγ × Eγ)
-
Calculate the projection data
P1 to
P9
-
Calculate the image values
(R1 to
R7)
using simple (unfiltered) backprojection
-
What is the average contrast between the bone and soft tissue
regions?
Use the following definition of contrast:
C= ( RBone − RS.T.)
/ RS.T.
- MRI Imaging
An MRI system for imaging Hydrogen
(of Gryomagnetic ratio 42.58 MHz/T )
has a main magnet with a Magnetic Field
B0 of 1.0 T. Gradient fields
in the slice selection and frequency encoding directions
are of strength 0.5 Gauss/cm. The maximum gradient field
in the phase encoding direction is 0.1 Gauss/cm.
An 90°−FID pulse sequence is being used, with a
slice selection pulse of 5 ms, a phase encoding pulse
of 500 µs, and a frequency encoding pulse of 50 ms.
The 90°−FID signal is given by:
signal = k ρ ( 1 − exp(−TR/T1) )
-
What is the width of the selected slice?
Assume that the Fourier Transform of an RF pulse
of duration T has a frequency domain width of 1/T.
-
The clinical staff are imaging a part of the patients
brain. The field of view is desired to be 5 cm (In both
phase and frequency encoding directions)
What is
the maximum phase difference between the tissues
at opposite edges of the field of view?
-
What is the frequency encoding signal bandwidth
(from tissues in the field of view)?
-
The clinical staff are concerned that the patient may
have a particular type of cancer, which has a
T1 of 0.8 s, and density of 1.0 g/cm3.
Assume normal brain tissue has a
T1 of 1.2 s, and density of 0.95 g/cm3.
What is best choice of TR to
optimize contrast between that cancerous region
and the normal brain tissue?
-
Given this choice of TR, and the
encoding timings given previously, how many
slices can be imaged within the TR
interval?
-
The clinical staff wish to image 15 slices of tissue
with a resolution of 1 mm in both the frequency and phase
encoding directions.
How many RF pulse sequences will be required?
Using the value calculated in part E, how long will
this take?
- Nuclear Medical Imaging
-
In class we have discussed three kinds of collimation:
1) Pinhole collimation
2) Electronic collimation
3) Parallel hole collimation.
Discuss each collimation technology, describing
one medical imaging application, and describing
any limitations or advantages in terms of efficiency, focus,
and spatial uniformity.
-
Discuss two issues (of the three presented in class)
to be considered in the choice of isotopes
for nuclear medical imaging.
- Ultrasound
A circular ultrasound transducer of diameter 1 cm is being
used
at a pulse frequency of 1 MHz
to image the mitral valve of the heart (mode M imaging).
The background tissue
(ie. all tissue except the valve)
is a mixture of muscle, fat, and blood, with the
following average ultrasound parameters:
velocity= 1500 m/s,
attenuation= 1.5 db/cm, density = 0.95 g/cm3.
The mitral valve itself has the following parameters:
velocity= 1550 m/s,
attenuation= 1.8 db/cm, density = 1.00 g/cm3.
Use the weakly reflecting (ie. no reverberations) assumption.
-
We wish to image a mitral valve at a depth of
6 cm.
Is this depth within the near field for this
transducer?
-
What is the minimum pulse length (in µs),
such that the pulse length is two times greater
than the diffraction limit (ie. such that the
received signal at the transducer centre and
edge are within the pulse envelope) ?
-
Consider the body to be 30 cm across.
How rapidly can the pulse be repeated (scan rate), in order
to avoid contamination with the previous echo
signal?
At its maximum speed, consider the mitral valve to
move 1 cm in 50 ms.
How far will the mitral valve move between scans at
this maximum rate?
-
The uncertainty (position estimation error) in the
depth of the mitral valve has a contribution
from the movement, and from the from the pulse length.
Calculate the speed of movement of the mitral valve, at which
the uncertainty contribution from the movement and pulse
lengths are equal.
-
What is the reflectivity of the interface
between the mitral valve and background tissue?
-
Two separate echo signals are obtained when the mitral
valve is at a depth of 5 cm and 6 cm, respectively.
What is the ratio of pulse envelope amplitude between
the two echos from the mitral valve?
Last Updated:
$Date: 2005-03-03 16:46:17 -0500 (Thu, 03 Mar 2005) $
|


 Engineering
Engineering
 SCE
SCE
 Faculty
Faculty
 A. Adler
A. Adler
 Courses
Courses
 ELG7173
ELG7173
 Exam 2003
Exam 2003